merlin
The Metabolic Models Reconstruction Using Genome-Scale Information (merlin) tool is an user-friendly Java application that assists the reconstruction of genome-scale metabolic models for any organism that has its genome sequenced.
It performs the major steps of the reconstruction process, including the functional genomic annotation of the whole genome and subsequent construction of the portfolio of reactions. Moreover, merlin includes tools for the identification and annotation of genes encoding transport proteins, generating the transport reactions for those carriers. It also performs the compartmentalisation of the model, predicting the organelle localisation of the proteins encoded in the genome, and thus the localisation of the metabolites involved in the reactions promoted by such enzymes. The gene to proteins to reactions (GPR) associations are automatically generated and included in the model. Finally, merlin expedites the transition from genome-scale reconstructions to draft metabolic models exported in the SBML standard format, allowing the user to have a preliminary view of the biochemical network.
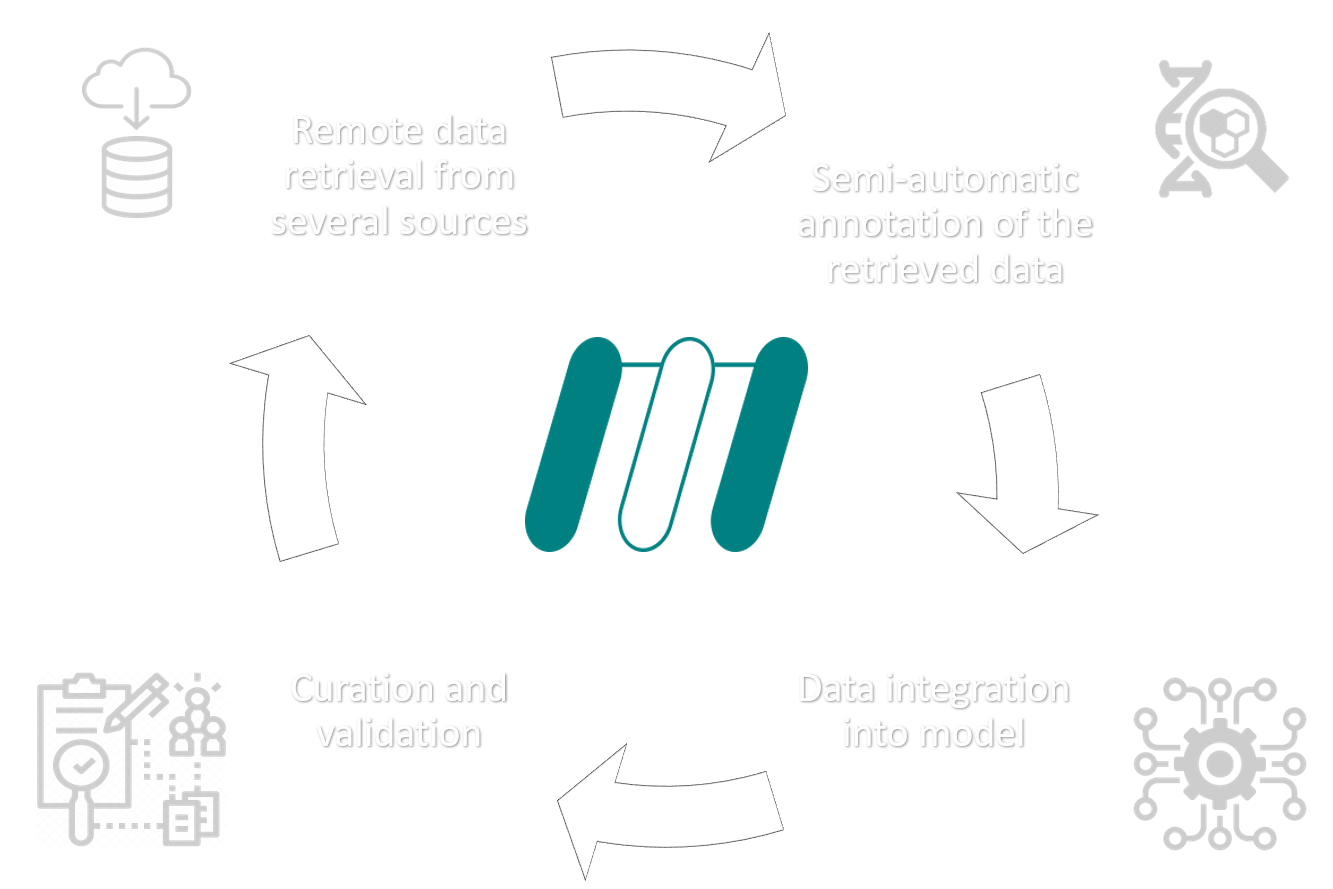

merlin makes it easier to reconstruct genome-scale metabolic models.
TranSyT
Transport reactions are added to genome-scale metabolic models based on experimental data and literature using the Transport Systems Tracker (TranSyT).
TranSyT is a tool to identify transport systems and the compounds carried across membranes, based on the annotations of the Transporter Classification Database (TCDB). In addition, this tool also generates the respective transport reactions while providing the respective Gene-Protein-Reaction associations.
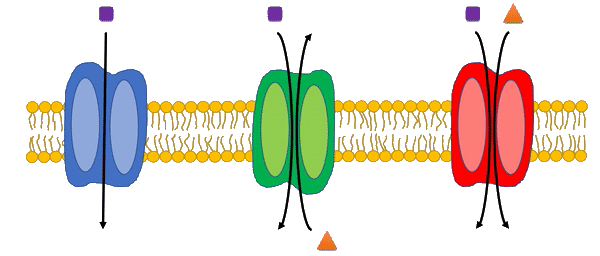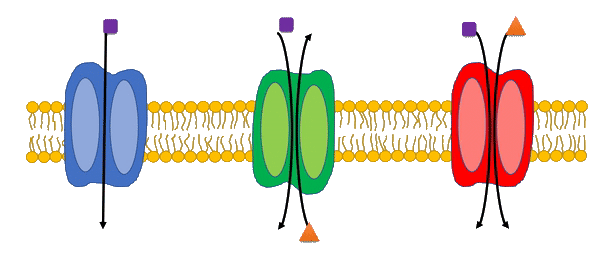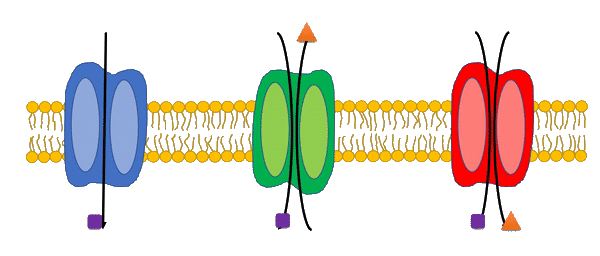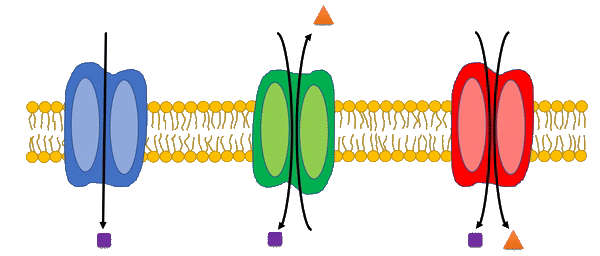
BioISO
Debugging and gap-finding genome-scale metabolic models using the Biological networks constraint-based In Silico Optimization (BioISO).
BioISO is aimed at evaluating either the biomass or metabolic network formulation of a given constraint-based metabolic model. This computational tool is based on the Constraints-Based Reconstruction and Analysis (COBRA) and Flux Balance Analysis (FBA) frameworks. BioISO can receive as input a constraint-based metabolic model in the SBML file format and return either potential errors in the biomass formulation or gaps in the metabolic network.